Download Certified Machine Learning Associate.CERTIFIED-MACHINE-LEARNING-ASSOCIATE.Pass4Success.2026-01-28.33q.vcex
| Vendor: | Databricks |
| Exam Code: | CERTIFIED-MACHINE-LEARNING-ASSOCIATE |
| Exam Name: | Certified Machine Learning Associate |
| Date: | Jan 28, 2026 |
| File Size: | 892 KB |
| Downloads: | 1 |
How to open VCEX files?
Files with VCEX extension can be opened by ProfExam Simulator.
Discount: 20%
Demo Questions
Question 1
A data scientist has defined a Pandas UDF function predict to parallelize the inference process for a single-node model:

They have written the following incomplete code block to use predict to score each record of Spark DataFrame spark_df:

Which of the following lines of code can be used to complete the code block to successfully complete the task?
- predict(*spark_df.columns)
- mapInPandas(predict)
- predict(Iterator(spark_df))
- mapInPandas(predict(spark_df.columns))
- predict(spark_df.columns)
Correct answer: B
Explanation:
To apply the Pandas UDF predict to each record of a Spark DataFrame, you use the mapInPandas method. This method allows the Pandas UDF to operate on partitions of the DataFrame as pandas DataFrames, applying the specified function (predict in this case) to each partition. The correct code completion to execute this is simply mapInPandas(predict), which specifies the UDF to use without additional arguments or incorrect function calls. Reference:PySpark DataFrame documentation (Using mapInPandas with UDFs). To apply the Pandas UDF predict to each record of a Spark DataFrame, you use the mapInPandas method. This method allows the Pandas UDF to operate on partitions of the DataFrame as pandas DataFrames, applying the specified function (predict in this case) to each partition. The correct code completion to execute this is simply mapInPandas(predict), which specifies the UDF to use without additional arguments or incorrect function calls. Reference:
PySpark DataFrame documentation (Using mapInPandas with UDFs).
Question 2
A data scientist has developed a machine learning pipeline with a static input data set using Spark ML, but the pipeline is taking too long to process. They increase the number of workers in the cluster to get the pipeline to run more efficiently. They notice that the number of rows in the training set after reconfiguring the cluster is different from the number of rows in the training set prior to reconfiguring the cluster.
Which of the following approaches will guarantee a reproducible training and test set for each model?
- Manually configure the cluster
- Write out the split data sets to persistent storage
- Set a speed in the data splitting operation
- Manually partition the input data
Correct answer: B
Explanation:
To ensure reproducible training and test sets, writing the split data sets to persistent storage is a reliable approach. This allows you to consistently load the same training and test data for each model run, regardless of cluster reconfiguration or other changes in the environment.Correct approach:Split the data.Write the split data to persistent storage (e.g., HDFS, S3).Load the data from storage for each model training session.train_df, test_df = spark_df.randomSplit([0.8, 0.2], seed=42) train_df.write.parquet('path/to/train_df.parquet') test_df.write.parquet('path/to/test_df.parquet') # Later, load the data train_df = spark.read.parquet('path/to/train_df.parquet') test_df = spark.read.parquet('path/to/test_df.parquet')Spark DataFrameWriter Documentation To ensure reproducible training and test sets, writing the split data sets to persistent storage is a reliable approach. This allows you to consistently load the same training and test data for each model run, regardless of cluster reconfiguration or other changes in the environment.
Correct approach:
Split the data.
Write the split data to persistent storage (e.g., HDFS, S3).
Load the data from storage for each model training session.
train_df, test_df = spark_df.randomSplit([0.8, 0.2], seed=42) train_df.write.parquet('path/to/train_df.parquet') test_df.write.parquet('path/to/test_df.parquet') # Later, load the data train_df = spark.read.parquet('path/to/train_df.parquet') test_df = spark.read.parquet('path/to/test_df.parquet')
Spark DataFrameWriter Documentation
Question 3
Which statement describes a Spark ML transformer?
- A transformer is an algorithm which can transform one DataFrame into another DataFrame
- A transformer is a hyperparameter grid that can be used to train a model
- A transformer chains multiple algorithms together to transform an ML workflow
- A transformer is a learning algorithm that can use a DataFrame to train a model
Correct answer: A
Explanation:
In Spark ML, a transformer is an algorithm that can transform one DataFrame into another DataFrame. It takes a DataFrame as input and produces a new DataFrame as output. This transformation can involve adding new columns, modifying existing ones, or applying feature transformations. Examples of transformers in Spark MLlib include feature transformers like StringIndexer, VectorAssembler, and StandardScaler.Databricks documentation on transformers: Transformers in Spark ML In Spark ML, a transformer is an algorithm that can transform one DataFrame into another DataFrame. It takes a DataFrame as input and produces a new DataFrame as output. This transformation can involve adding new columns, modifying existing ones, or applying feature transformations. Examples of transformers in Spark MLlib include feature transformers like StringIndexer, VectorAssembler, and StandardScaler.
Databricks documentation on transformers: Transformers in Spark ML
Question 4
A machine learning engineer would like to develop a linear regression model with Spark ML to predict the price of a hotel room. They are using the Spark DataFrame train_df to train the model.
The Spark DataFrame train_df has the following schema:

The machine learning engineer shares the following code block:
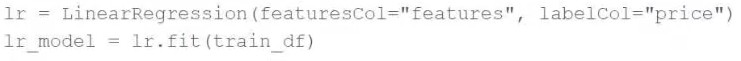
Which of the following changes does the machine learning engineer need to make to complete the task?
- They need to call the transform method on train df
- They need to convert the features column to be a vector
- They do not need to make any changes
- They need to utilize a Pipeline to fit the model
- They need to split the features column out into one column for each feature
Correct answer: B
Explanation:
In Spark ML, the linear regression model expects the feature column to be a vector type. However, if the features column in the DataFrame train_df is not already in this format (such as being a column of type UDT or a non-vectorized type), the engineer needs to convert it to a vector column using a transformer like VectorAssembler. This is a critical step in preparing the data for modeling as Spark ML models require input features to be combined into a single vector column.ReferenceSpark MLlib documentation for LinearRegression: https://spark.apache.org/docs/latest/ml-classification-regression.html#linear-regression In Spark ML, the linear regression model expects the feature column to be a vector type. However, if the features column in the DataFrame train_df is not already in this format (such as being a column of type UDT or a non-vectorized type), the engineer needs to convert it to a vector column using a transformer like VectorAssembler. This is a critical step in preparing the data for modeling as Spark ML models require input features to be combined into a single vector column.
Reference
Spark MLlib documentation for LinearRegression: https://spark.apache.org/docs/latest/ml-classification-regression.html#linear-regression
Question 5
A data scientist wants to efficiently tune the hyperparameters of a scikit-learn model. They elect to use the Hyperopt library's fmin operation to facilitate this process. Unfortunately, the final model is not very accurate. The data scientist suspects that there is an issue with the objective_function being passed as an argument to fmin.
They use the following code block to create the objective_function:

Which of the following changes does the data scientist need to make to their objective_function in order to produce a more accurate model?
- Add test set validation process
- Add a random_state argument to the RandomForestRegressor operation
- Remove the mean operation that is wrapping the cross_val_score operation
- Replace the r2 return value with -r2
- Replace the fmin operation with the fmax operation
Correct answer: D
Explanation:
When using the Hyperopt library with fmin, the goal is to find the minimum of the objective function. Since you are using cross_val_score to calculate the R2 score which is a measure of the proportion of the variance for a dependent variable that's explained by an independent variable(s) in a regression model, higher values are better. However, fmin seeks to minimize the objective function, so to align with fmin's goal, you should return the negative of the R2 score (-r2). This way, by minimizing the negative R2, fmin is effectively maximizing the R2 score, which can lead to a more accurate model.ReferenceHyperopt Documentation: http://hyperopt.github.io/hyperopt/Scikit-Learn documentation on model evaluation: https://scikit-learn.org/stable/modules/model_evaluation.html When using the Hyperopt library with fmin, the goal is to find the minimum of the objective function. Since you are using cross_val_score to calculate the R2 score which is a measure of the proportion of the variance for a dependent variable that's explained by an independent variable(s) in a regression model, higher values are better. However, fmin seeks to minimize the objective function, so to align with fmin's goal, you should return the negative of the R2 score (-r2). This way, by minimizing the negative R2, fmin is effectively maximizing the R2 score, which can lead to a more accurate model.
Reference
Hyperopt Documentation: http://hyperopt.github.io/hyperopt/
Scikit-Learn documentation on model evaluation: https://scikit-learn.org/stable/modules/model_evaluation.html
Question 6
A data scientist is developing a single-node machine learning model. They have a large number of model configurations to test as a part of their experiment. As a result, the model tuning process takes too long to complete. Which of the following approaches can be used to speed up the model tuning process?
- Implement MLflow Experiment Tracking
- Scale up with Spark ML
- Enable autoscaling clusters
- Parallelize with Hyperopt
Correct answer: D
Explanation:
To speed up the model tuning process when dealing with a large number of model configurations, parallelizing the hyperparameter search using Hyperopt is an effective approach. Hyperopt provides tools like SparkTrials which can run hyperparameter optimization in parallel across a Spark cluster.Example:from hyperopt import fmin, tpe, hp, SparkTrials search_space = { 'x': hp.uniform('x', 0, 1), 'y': hp.uniform('y', 0, 1) } def objective(params): return params['x'] ** 2 + params['y'] ** 2 spark_trials = SparkTrials(parallelism=4) best = fmin(fn=objective, space=search_space, algo=tpe.suggest, max_evals=100, trials=spark_trials)Hyperopt Documentation To speed up the model tuning process when dealing with a large number of model configurations, parallelizing the hyperparameter search using Hyperopt is an effective approach. Hyperopt provides tools like SparkTrials which can run hyperparameter optimization in parallel across a Spark cluster.
Example:
from hyperopt import fmin, tpe, hp, SparkTrials search_space = { 'x': hp.uniform('x', 0, 1), 'y': hp.uniform('y', 0, 1) } def objective(params): return params['x'] ** 2 + params['y'] ** 2 spark_trials = SparkTrials(parallelism=4) best = fmin(fn=objective, space=search_space, algo=tpe.suggest, max_evals=100, trials=spark_trials)
Hyperopt Documentation
Question 7
A data scientist wants to tune a set of hyperparameters for a machine learning model. They have wrapped a Spark ML model in the objective function objective_function and they have defined the search space search_space.
As a result, they have the following code block:

Which of the following changes do they need to make to the above code block in order to accomplish the task?
- Change SparkTrials() to Trials()
- Reduce num_evals to be less than 10
- Change fmin() to fmax()
- Remove the trials=trials argument
- Remove the algo=tpe.suggest argument
Correct answer: A
Explanation:
The SparkTrials() is used to distribute trials of hyperparameter tuning across a Spark cluster. If the environment does not support Spark or if the user prefers not to use distributed computing for this purpose, switching to Trials() would be appropriate. Trials() is the standard class for managing search trials in Hyperopt but does not distribute the computation. If the user is encountering issues with SparkTrials() possibly due to an unsupported configuration or an error in the cluster setup, using Trials() can be a suitable change for running the optimization locally or in a non-distributed manner.ReferenceHyperopt documentation: http://hyperopt.github.io/hyperopt/ The SparkTrials() is used to distribute trials of hyperparameter tuning across a Spark cluster. If the environment does not support Spark or if the user prefers not to use distributed computing for this purpose, switching to Trials() would be appropriate. Trials() is the standard class for managing search trials in Hyperopt but does not distribute the computation. If the user is encountering issues with SparkTrials() possibly due to an unsupported configuration or an error in the cluster setup, using Trials() can be a suitable change for running the optimization locally or in a non-distributed manner.
Reference
Hyperopt documentation: http://hyperopt.github.io/hyperopt/
Question 8
What is the name of the method that transforms categorical features into a series of binary indicator feature variables?
- Leave-one-out encoding
- Target encoding
- One-hot encoding
- Categorical
- String indexing
Correct answer: C
Explanation:
The method that transforms categorical features into a series of binary indicator variables is known as one-hot encoding. This technique converts each categorical value into a new binary column, which is essential for models that require numerical input. One-hot encoding is widely used because it helps to handle categorical data without introducing a false ordinal relationship among categories. Reference:Feature Engineering Techniques (One-Hot Encoding). The method that transforms categorical features into a series of binary indicator variables is known as one-hot encoding. This technique converts each categorical value into a new binary column, which is essential for models that require numerical input. One-hot encoding is widely used because it helps to handle categorical data without introducing a false ordinal relationship among categories. Reference:
Feature Engineering Techniques (One-Hot Encoding).
Question 9
A machine learning engineer has created a Feature Table new_table using Feature Store Client fs. When creating the table, they specified a metadata description with key information about the Feature Table. They now want to retrieve that metadata programmatically.
Which of the following lines of code will return the metadata description?
- There is no way to return the metadata description programmatically.
- fs.create_training_set('new_table')
- fs.get_table('new_table').description
- fs.get_table('new_table').load_df()
- fs.get_table('new_table')
Correct answer: C
Explanation:
To retrieve the metadata description of a feature table created using the Feature Store Client (referred here as fs), the correct method involves calling get_table on the fs client with the table name as an argument, followed by accessing the description attribute of the returned object. The code snippet fs.get_table('new_table').description correctly achieves this by fetching the table object for 'new_table' and then accessing its description attribute, where the metadata is stored. The other options do not correctly focus on retrieving the metadata description. Reference:Databricks Feature Store documentation (Accessing Feature Table Metadata). To retrieve the metadata description of a feature table created using the Feature Store Client (referred here as fs), the correct method involves calling get_table on the fs client with the table name as an argument, followed by accessing the description attribute of the returned object. The code snippet fs.get_table('new_table').description correctly achieves this by fetching the table object for 'new_table' and then accessing its description attribute, where the metadata is stored. The other options do not correctly focus on retrieving the metadata description. Reference:
Databricks Feature Store documentation (Accessing Feature Table Metadata).
Question 10
Which of the following is a benefit of using vectorized pandas UDFs instead of standard PySpark UDFs?
- The vectorized pandas UDFs allow for the use of type hints
- The vectorized pandas UDFs process data in batches rather than one row at a time
- The vectorized pandas UDFs allow for pandas API use inside of the function
- The vectorized pandas UDFs work on distributed DataFrames
- The vectorized pandas UDFs process data in memory rather than spilling to disk
Correct answer: B
Explanation:
Vectorized pandas UDFs, also known as Pandas UDFs, are a powerful feature in PySpark that allows for more efficient operations than standard UDFs. They operate by processing data in batches, utilizing vectorized operations that leverage pandas to perform operations on whole batches of data at once. This approach is much more efficient than processing data row by row as is typical with standard PySpark UDFs, which can significantly speed up the computation.ReferencePySpark Documentation on UDFs: https://spark.apache.org/docs/latest/api/python/user_guide/sql/arrow_pandas.html#pandas-udfs-a-k-a-vectorized-udfs Vectorized pandas UDFs, also known as Pandas UDFs, are a powerful feature in PySpark that allows for more efficient operations than standard UDFs. They operate by processing data in batches, utilizing vectorized operations that leverage pandas to perform operations on whole batches of data at once. This approach is much more efficient than processing data row by row as is typical with standard PySpark UDFs, which can significantly speed up the computation.
Reference
PySpark Documentation on UDFs: https://spark.apache.org/docs/latest/api/python/user_guide/sql/arrow_pandas.html#pandas-udfs-a-k-a-vectorized-udfs
Question 11
A data scientist wants to parallelize the training of trees in a gradient boosted tree to speed up the training process. A colleague suggests that parallelizing a boosted tree algorithm can be difficult.
Which of the following describes why?
- Gradient boosting is not a linear algebra-based algorithm which is required for parallelization
- Gradient boosting requires access to all data at once which cannot happen during parallelization.
- Gradient boosting calculates gradients in evaluation metrics using all cores which prevents parallelization.
- Gradient boosting is an iterative algorithm that requires information from the previous iteration to perform the next step.
Correct answer: D
Explanation:
Gradient boosting is fundamentally an iterative algorithm where each new tree is built based on the errors of the previous ones. This sequential dependency makes it difficult to parallelize the training of trees in gradient boosting, as each step relies on the results from the preceding step. Parallelization in this context would undermine the core methodology of the algorithm, which depends on sequentially improving the model's performance with each iteration. Reference:Machine Learning Algorithms (Challenges with Parallelizing Gradient Boosting).Gradient boosting is an ensemble learning technique that builds models in a sequential manner. Each new model corrects the errors made by the previous ones. This sequential dependency means that each iteration requires the results of the previous iteration to make corrections. Here is a step-by-step explanation of why this makes parallelization challenging:Sequential Nature: Gradient boosting builds one tree at a time. Each tree is trained to correct the residual errors of the previous trees. This requires the model to complete one iteration before starting the next.Dependence on Previous Iterations: The gradient calculation at each step depends on the predictions made by the previous models. Therefore, the model must wait until the previous tree has been fully trained and evaluated before starting to train the next tree.Difficulty in Parallelization: Because of this dependency, it is challenging to parallelize the training process. Unlike algorithms that process data independently in each step (e.g., random forests), gradient boosting cannot easily distribute the work across multiple processors or cores for simultaneous execution.This iterative and dependent nature of the gradient boosting process makes it difficult to parallelize effectively.ReferenceGradient Boosting Machine Learning AlgorithmUnderstanding Gradient Boosting Machines Gradient boosting is fundamentally an iterative algorithm where each new tree is built based on the errors of the previous ones. This sequential dependency makes it difficult to parallelize the training of trees in gradient boosting, as each step relies on the results from the preceding step. Parallelization in this context would undermine the core methodology of the algorithm, which depends on sequentially improving the model's performance with each iteration. Reference:
Machine Learning Algorithms (Challenges with Parallelizing Gradient Boosting).
Gradient boosting is an ensemble learning technique that builds models in a sequential manner. Each new model corrects the errors made by the previous ones. This sequential dependency means that each iteration requires the results of the previous iteration to make corrections. Here is a step-by-step explanation of why this makes parallelization challenging:
Sequential Nature: Gradient boosting builds one tree at a time. Each tree is trained to correct the residual errors of the previous trees. This requires the model to complete one iteration before starting the next.
Dependence on Previous Iterations: The gradient calculation at each step depends on the predictions made by the previous models. Therefore, the model must wait until the previous tree has been fully trained and evaluated before starting to train the next tree.
Difficulty in Parallelization: Because of this dependency, it is challenging to parallelize the training process. Unlike algorithms that process data independently in each step (e.g., random forests), gradient boosting cannot easily distribute the work across multiple processors or cores for simultaneous execution.
This iterative and dependent nature of the gradient boosting process makes it difficult to parallelize effectively.
Reference
Gradient Boosting Machine Learning Algorithm
Understanding Gradient Boosting Machines
HOW TO OPEN VCE FILES
Use VCE Exam Simulator to open VCE files
HOW TO OPEN VCEX AND EXAM FILES
Use ProfExam Simulator to open VCEX and EXAM files
ProfExam at a 20% markdown
You have the opportunity to purchase ProfExam at a 20% reduced price
Get Now!